Biomolecular engineering to make epigenetic knowledge useful
We are working to develop new methods for detecting gene-specific DNA methylation that are amenable to clinical use.
Non-coding, epigenetic modifications to DNA, in addition to the underlying ATCG code, have been shown to regulate gene transcription and play an important role in diseases such as cancer. Recent studies indicate that one such modification in promoter regions, DNA methylation, has value as a predictive biomarker. Current clinical practice relies on chemical conversion via bisulfite treatment to distinguish between methylated and unmethylated DNA; such treatment can suffer from inaccurate results due to incomplete conversion and DNA degradation. Previously, we engineered a methyl CpG binding domain (MBD) protein which can bind specifically to hemimethylated target DNA. Using this engineered MBD we developed a magnetic bead-based assay that is capable of detecting methylated target DNA in picomolar range concentration. We are now exploring kinetic characteristics of different hemimethylated DNA binding proteins and modeling the assay kinetics in order to design an epigenotyping assay that can be used to accurately and sensitively detect the gene-specific methylation in the clinical setting.
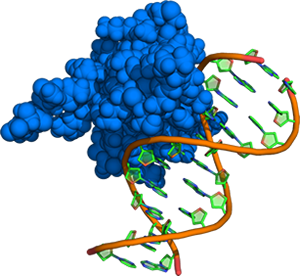
A methyl binding domain in complex with DNA bearing a symmetrically methylated CpG site. Methylation of cytosines in this sequence context in gene promoters leads to reduced expression of certain proteins in many cancers.